VIDEO and FREE TRIAL
Qlucore Omics Explorer video
Qlucore Omics Explorer free trial
Qlucore Diagnostics is a CE-marked IVDR software offering RNA-seq based cancer diagnostics. The first approved model is for Acute Lymphoblastic Leukemia (BCP-ALL). It includes both gene expression subtype classification of B-cell leukemias and gene fusion analysis support.
Key benefits:
Read our leaflet for a summary.
Qlucore Diagnostics BCP-ALL is a CE-marked IVDR software designed for the qualitative determination of clinically relevant genetic markers in samples from bone marrow or peripheral blood during the genetic workup of pediatric B-cell precursor Acute Lymphoblastic Leukemia (BCP-ALL).
The software supports RNA-Seq data analysis using gene expression-based classification and gene fusion identification. Classification is achieved through a machine learning-based classifier, providing probability scores for the following genetic subtypes:
Gene fusion identification is performed by fusion callers, with identified gene fusions exported to a report along with fusion breakpoints. The gene fusions are organized in three tiers based on documented clinical relevance. All gene fusions matching quality criteria are included to enable additional information on novel gene fusions.
The results aid in the initial classification and management of pediatric patients aged from one up to 18 years with suspected or diagnosed BCP-ALL. The Qlucore Diagnostics BCP-ALL is intended to be used with the Qlucore Diagnostics Platform software.
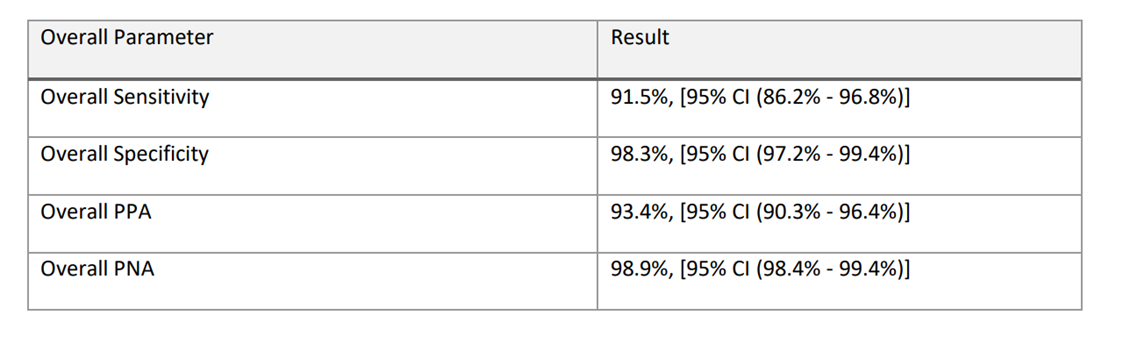
Clinical performance was evaluated on a representative sample of the intended population for the product in a multi-site study. Genetic subtype classification and gene fusion detection were performed by comparing the outcome of Qlucore Diagnostics BCP-ALL to the known status of the patient sample. The results of the clinical data were analyzed to produce sensitivity, specificity, percent positive agreement (PPA), percent negative agreement (PNA). The figure below presents the clinical performance for Qlucore Diagnostics BCP-ALL subtype classification.
The output of Qlucore Diagnostics is a pdf report that includes conclusions, results, plots, quality metrics and method information. Visualizations are included in the clinical report for better communication between a molecular genetics lab, clinicians and patients.
View a draft example of how a report might look.
White paper on RNA-sequencing as an emerging clinical diagnostic tool for BCP-ALL.
Based on Qlucore Insights, with research use only models for BCP-ALL, AML, lung and bladder cancer, there is a case study for BCP-ALL based on the use at Rigshospitalet in Copenhagen, enhancing their diagnostic capabilities.

