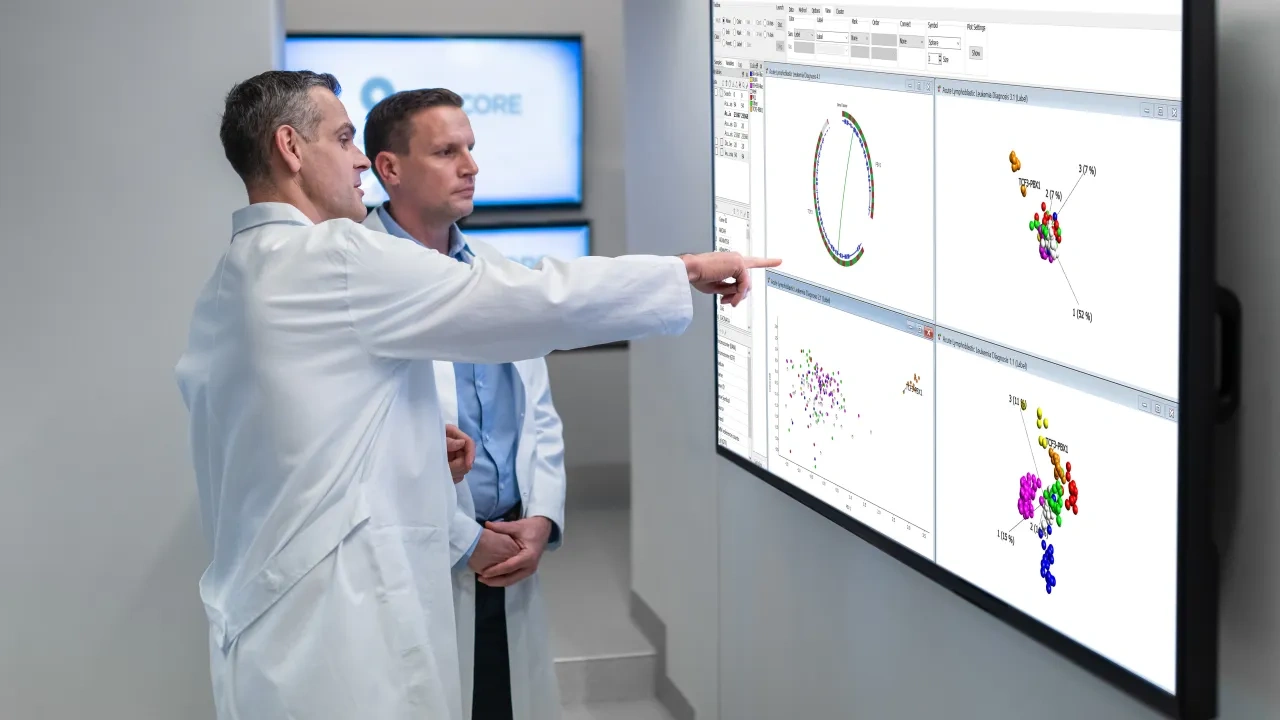
RNA-seq Analysis
Qlucore Omics Explorer is a next-generation bioinformatics software for research in life science, plant- and biotech industries, as well as academia. The easy to use visualization-based data analysis tool with inbuilt powerful statistics delivers immediate results and provides instant exploration and visualization of big data including rna-seq data.
Analysis of RNA-seq data
Bioinformatic software solutions and tools for analysis of RNA-seq (rnaseq) data tend to be complex. Qlucore Omics Explorer makes the analysis of RNA-seq data easy and accessible for biologists and bench scientists.
The main application is to work with digital gene expression. The inbuilt workflow for RNA-seq data includes a first step for import of aligned BAM files. The second step is normalization based on the VST, TMM, TPM or FPKM methods. After this the bioinformatic analysis and visualization of the RNA-seq data can start. A wide range of statistical methods such DESeq2 methods and ANOVA are available.
If your RNA-seq data has already been pre-normalized, you can import it in a txt of csv file using the Wizard.
All functionality in Qlucore Omics Explorer can be used to provide new and actionable insights.

Key functionalities:
- Import and normalize aligned BAM files
- Identify discriminating features(genes) with a few mouse clicks
- Visualize data and do data mining and data exploration
- Show results in the flexible and easy-to-use heatmap with hierarchical clustering
- Verify your hypotheses using powerful statistics including DESeq2 methods and ANOVA and different forms of regressions
- Compare with pathways and other biological information using the integrated and user friendly Gene Set Enrichment analysis (GSEA) Workbench
- No wait, all analysis, statistics and plots are updated directly
- Extensive plot library both in 2D and 3D - e.g PCA, heatmaps, UMAP, t-SNE, box, scatter, volcano plots, Venn diagram, Kaplan Meier plots etc.
- Use the Genome Browser to visualize your genes of interest
Read about single cell RNA-seq analysis.
Import RNA-seq data
Does it work on my data?
Answer the four quick questions below and find out if you can use Qlucore on your data.
For more details about supported data formats and data import see Data Import or Contact us with questions.
Case studies
RNA-Seq analysis using Qlucore
Stanford University, USPerforming gene expression analysis based on RNA sequencing data, in Dilated Cardiomyopathy studies.
Qlucore analysis of transcriptomic data
University of Manchester, UKThe study includes working with data from more than 400 arrays. Visualization is used in the effort to understand the human growth process.