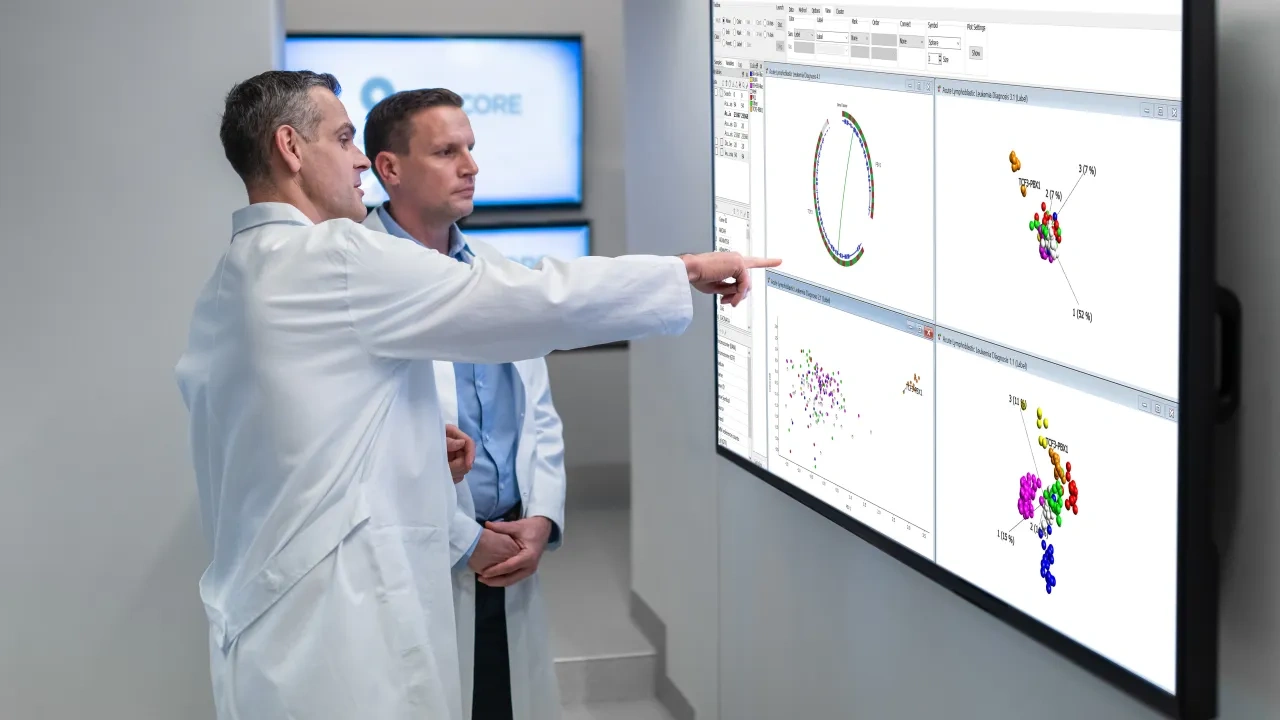
Qlucore Insights
AI-based disease subclassification
Advances in precision medicine require powerful yet accessible tools and methods that unite common goals with collaborative effort. Qlucore Insights is a powerful and flexible software platform that enables implementation of disease-specific tests (models) for subtype classification. Qlucore Insights is the starting point to use NGS based data in machine learning based classifiers and gene fusion identification. Further, researchers can use it to build and evaluate new models, or as the starting point for companion diagnostics. And frontline labs now have a powerful yet user-friendly tool for transforming clinical data into new insights, enabling new RNA-seq, transcriptomic-based or molecular-based research.
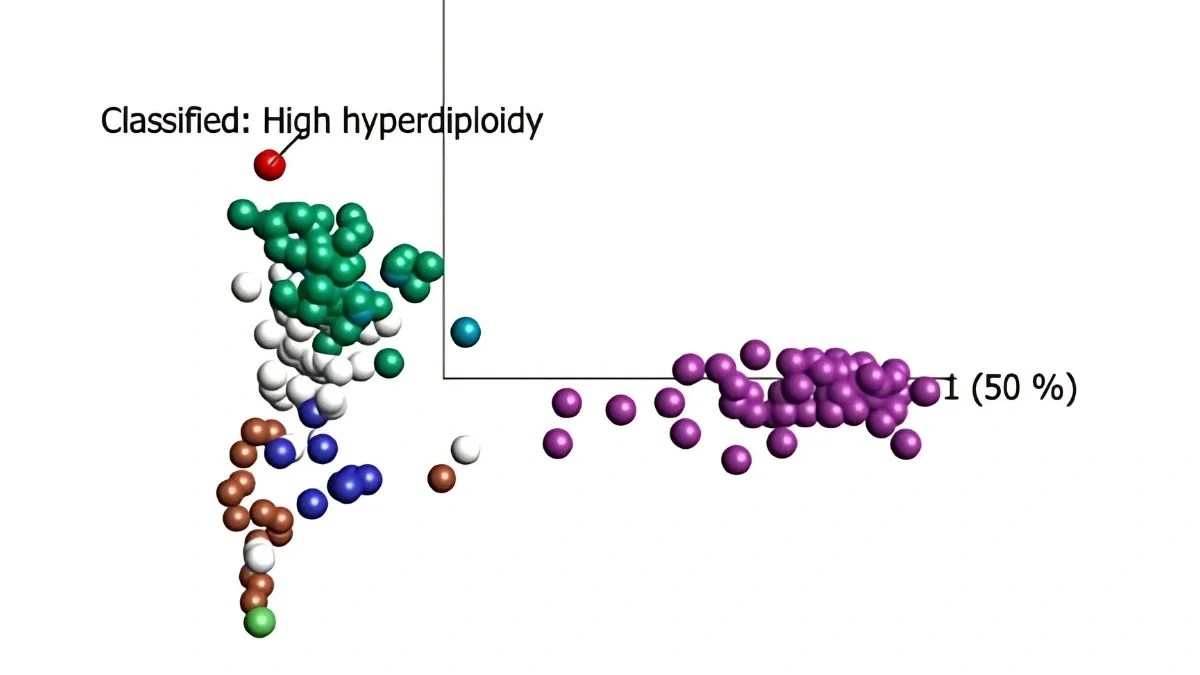
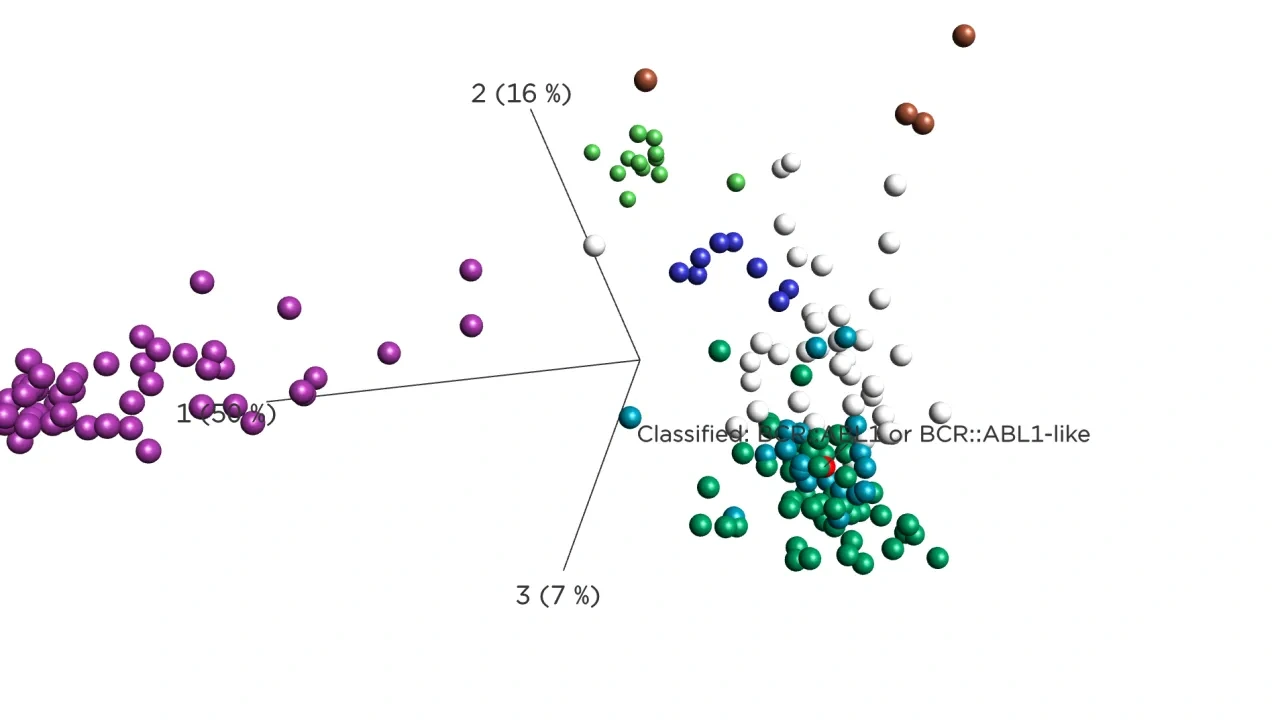
Rigshospitalet BCP-ALL Case
A team at Rigshospitalet, Copenhagen, Denmark has implemented the use of Qlucore Insights for in-house use, in accordance with Article 5(5) in EU IVDR, and included it in the RNA lab workflow for analysis of pediatric leukemia. As data input, whole RNA sequencing data (WTS) from diagnostic ALL samples were used.
Tests
There are currently four tests available, using both gene expression subtyping and gene fusion analysis.
Pediatric Leukemia (BCP-ALL)
Acute Myleoid Leukemia (AML)
Lung Cancer
Bladder Cancer
Test details
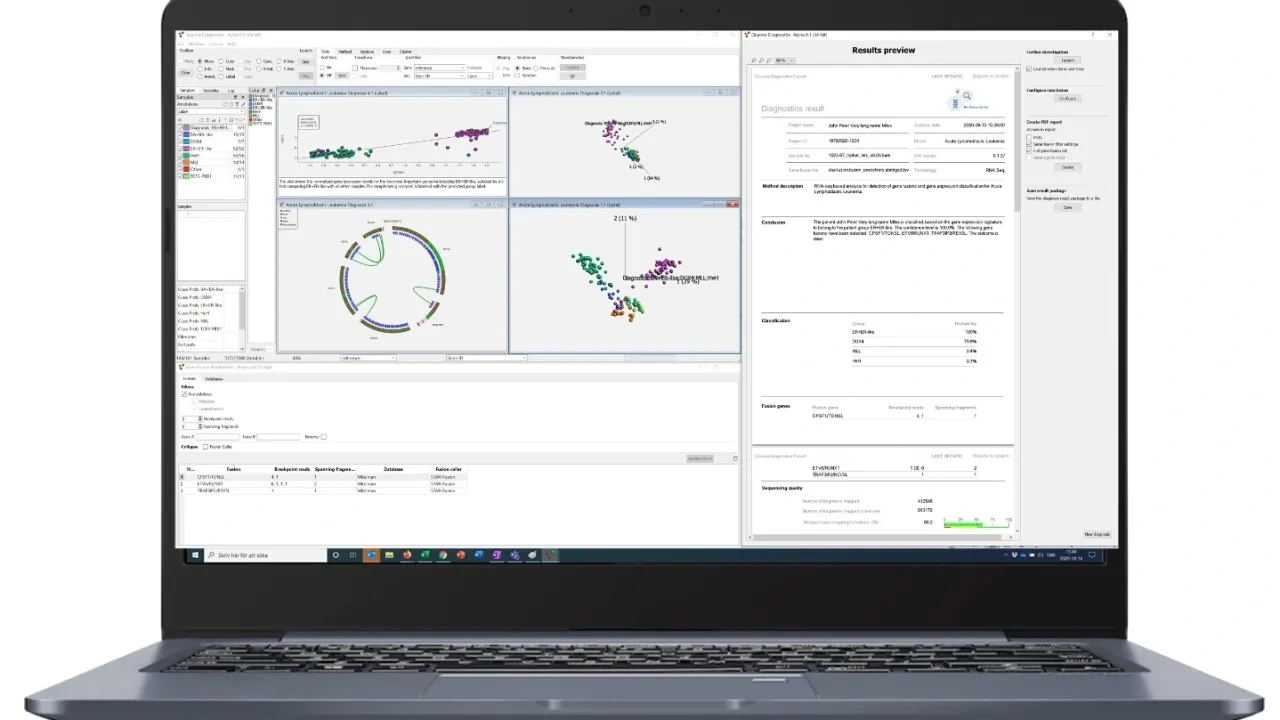
RNA-seq analysis software for better insights
Qlucore Insights enables disease specific subtype classification and first-class gene fusion analysis support of individual samples.
Qlucore Insights is an easy-to-use software that takes RNA-seq data as input to generate a report with subtype probabilities. The classification is done using a machine learning classifier with gene expression levels as input. The report also includes identified gene fusions. The selection of gene fusions is generated using flexible filter settings. Qlucore Insights is a flexible platform that allows users to choose from a range of disease-specific models for tailored analysis. The tests (models) plug into the Qlucore Insight platform and create one unified solution for the lab to manage a range of tumor types. The wet lab workflow is similar for all models and it is based on Illumina RNA-seq kits. The bioinformatic pipeline uses open-source tools.
Visual reporting
The subtype classification is complemented with flexible visualizations which put the sample in context with reference data by using well-characterized disease cases. Complementing the reports with easy-to-interpret visualizations improves the analysis and communication.
Videos
Introduction
Qlucore Insights for BCP-ALL
Qlucore Insights is our software for clinical data analysis. It supports a broad range of tests. This demonstration focus on pediatric leukemia. The t...
Watch now
Introduction
Qlucore Insights for Lung Cancer
Qlucore Insights is our software for clinical data analysis. It supports a broad range of tests. This demonstration focuses on lung cancer lesions. Th...
Watch now
Introduction
Qlucore Insights
A product transforming clinical data into new insights to aid clinicians and lab personnel in their work with precision medicine.The program supports ...
Watch now
How it works
The program includes AI powered subtype classification of individual samples, with flexible visualization and comprehensive reporting. Additionally the user-friendly interface for exploration of detected gene fusions and database integration provides a future proof solution adapting to new medical findings.