News
Here you will find all the newsletters. If you would like to receive newsletters automatically, create an account and accept to receive newsletters. Create an account
Version 3.11 is launched
Qlucore Omics Explorer (QOE) version 3.11 is launched. This release places special focus on tools and functionality that make proteomics analysis easi...
Read more

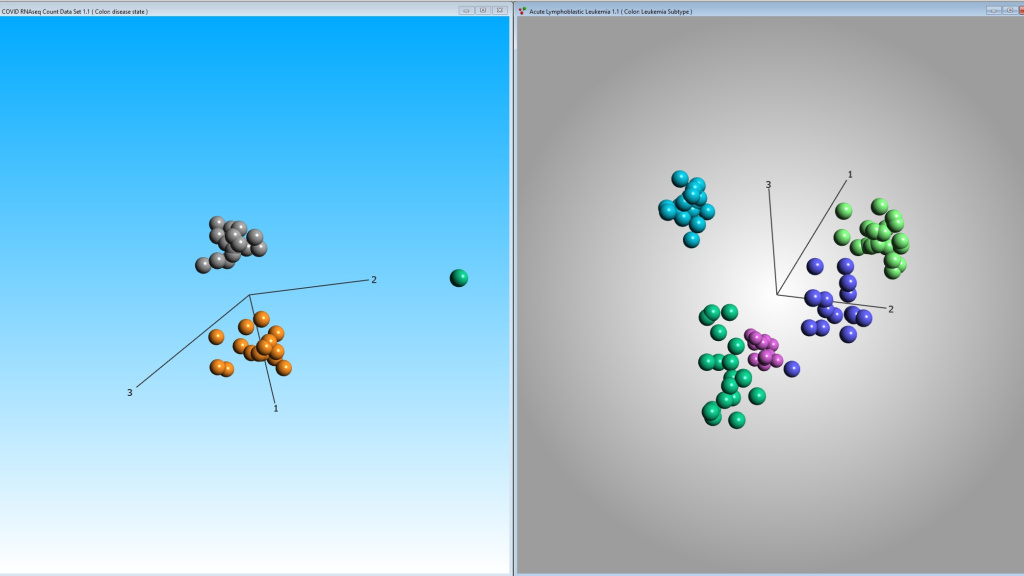
Qlucore Newsletter: RNA Sequencing is Transforming Diagnostics
RNA sequencing (RNA-seq) is rapidly proving its value in diagnostics, offering insights into gene expression, fusions, and mutations.
Read more
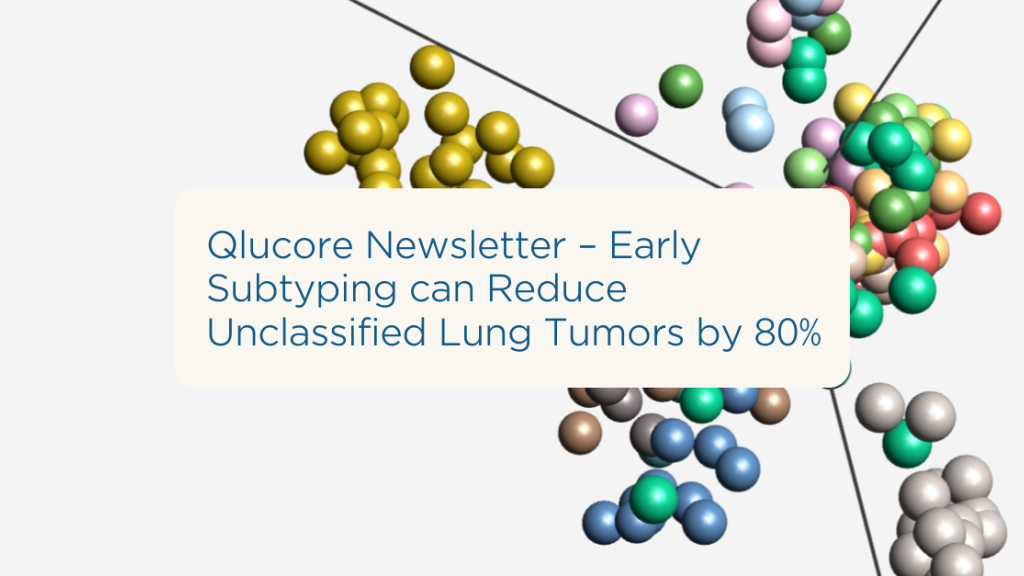
Qlucore Newsletter: Early Subtyping can Reduce Unclassified Lung Tumors by 80%
Unclassified tumors delay analysis and decision-making.
Read more
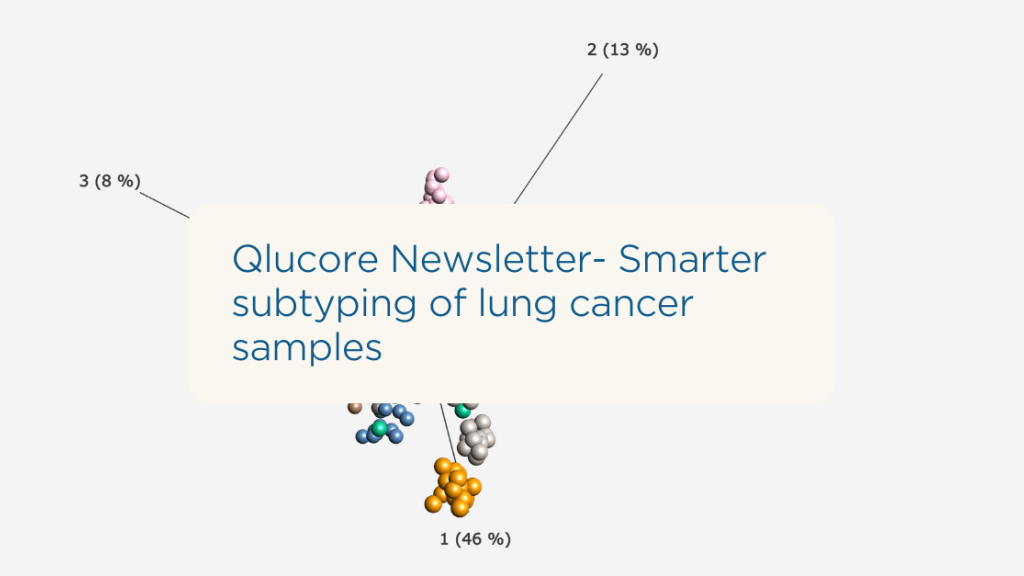
Qlucore Newsletter: Smarter Subtyping of Lung Cancer Samples
Looking for smarter lung cancer sample analysis?
Read more

Qlucore Newsletter: Find Patterns and Insights in Your Data
Heatmaps are powerful visualizations. They can help you uncover hidden patterns and insights in your data.
Read more

Qlucore Newsletter: Free Trainings – Available at some of Europe´s leading Research Hubs
Get better answers from your data. Enhance your understanding and use of Qlucore Omics Explorer.
Read more

Qlucore Newsletter: Let Your Data Speak with Clarity and Impact
Thousands of researchers have already benefited from the easy-to-use Qlucore Omics Explorer software, to accelerate their path to publication. Are you...
Read more

Use RNA-Seq to Sharpen Diagnostic Precision
RNA sequencing (RNA-seq) can be used to measure gene expression patterns and gene fusions, crucial for accurate diagnosis and treatment planning. ...
Read more

Qlucore Newsletter: The Vital Role of RNA Sequencing in Modern Diagnostics
A recent paper* published in the Lancet discovery science suite of journals underscores the importance of RNA sequencing (RNA-seq) in the field of dia...
Read more

Qlucore Newsletter: The Power of Transcriptomics in Cancer Research
In the rapidly changing field of cancer research, understanding molecular dynamics is crucial.
Read more

Qlucore Newsletter: IVDR Test for Pediatric Leukemia Now in Clinical Use
Rigshospitalet, Denmark's largest teaching and public hospital, uses Qlucore Diagnostics BCP-ALL.
Read more

Qlucore Newsletter: New Version of Qlucore Omics Explorer in the Cloud
Run an analysis and share it in a moment regardless of where you are?