Acute Lymphoblastic Leukemia (BCP-ALL) model (RuO)
Acute lymphoblastic leukemia (ALL) is the most common acute leukemia in children and accounts for approximately 30% of all cases of acute leukemia in adults. This disease is characterized by several genetic alterations of great clinical diagnostic importance, as recognized by the current WHO classification.
Contact us to find out more
Routine Diagnostics of ALL
Today more than four different methods (e.g. chromosome banding analysis, genetic assays identifying SNVs and CNVs, FISH) are used in routine diagnostics for diagnosis of ALL. These requires several different competences, instruments and are collectively time consuming.
A number of gene fusions in ALL can be targeted with specific drugs with improved outcomes for patients, or used for risk stratification, making it critical to detect such alterations at the diagnostic workup. In this context, RNA-sequencing is becoming an increasingly important diagnostic tool as it can detect gene fusions in an unbiased manner and aid the subtype classification based on distinct gene expression signatures.
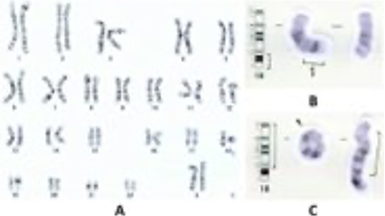
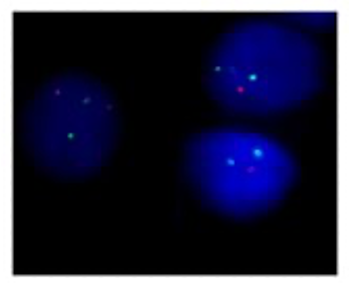
RNA-seq - emerging clinical diagnostic tool for ALL
RNA sequencing allows measuring of two key parameters for diagnostics of Acute Lymphoblastic Leukemia which can be used for subtype classification:
1) chimeric gene fusions
2) gene expression levels
Given high resolution enables the detection of most clinically relevant genetic alterations, the decreasing sequencing costs associated with such tests, and amenability to automatization we will see a wide adoption of RNA-sequencing in the clinical setting with several frontline laboratories already offering such analyses.
Read more about the power of integrating transcriptomics in diagnostics and precision medicine:
"Transcriptomics paving the way for improved diagnostics and precision medicine of acute leukemia" Fioretos et al.
Performance (RoU)
The Qlucore Insights gene expression classifier model for pediatric Acute Lymphoblastic Leukemia (BCP-ALL) allows gene expression based subtype classification of the B-cell leukemias listed below, and with the gene fusions analysis support further stratification is possible. For research use only (RoU).
1. High hyperdiploidy*
2. ETV6::RUNX1* + ETV6::RUNX1-like
3. KMT2A(MLL)-rearranged (MLL)*
4. TCF3::PBX1*
5. BCR::ABL1* + BCR::ABL1-like*
6. DUX4-rearranged
*RECOGNIZED BY THE CURRENT WHO CLASSIFICATION
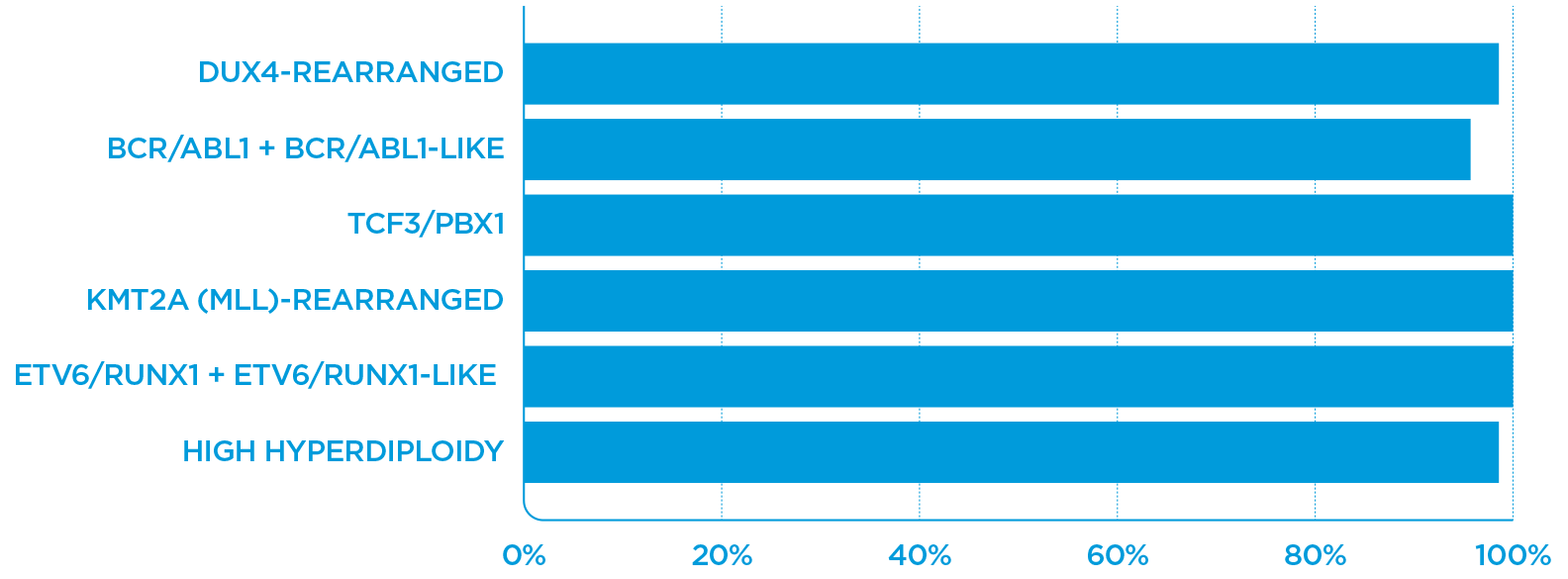
The performance plot illustrates the accuracy of the gene expression classification from validation with external data. Note: These results are preliminary and may change.
White paper - Genetic classification of ALL
RNA-sequencing as an emerging clinical diagnostic tool. READ WHITE PAPER
Poster - presented at AACR special conference in cancer research - Gene expression-based classifier for pediatric BCP-ALL subtypes
Poster presented at AACR special conference in cancer research: advances in pediatric cancer research, September 5-8, 2024, Toronto, Ontario, Canada. Cancer Res (2024) 84 (17_Supplement): B027.