Metabolomics data analysis software
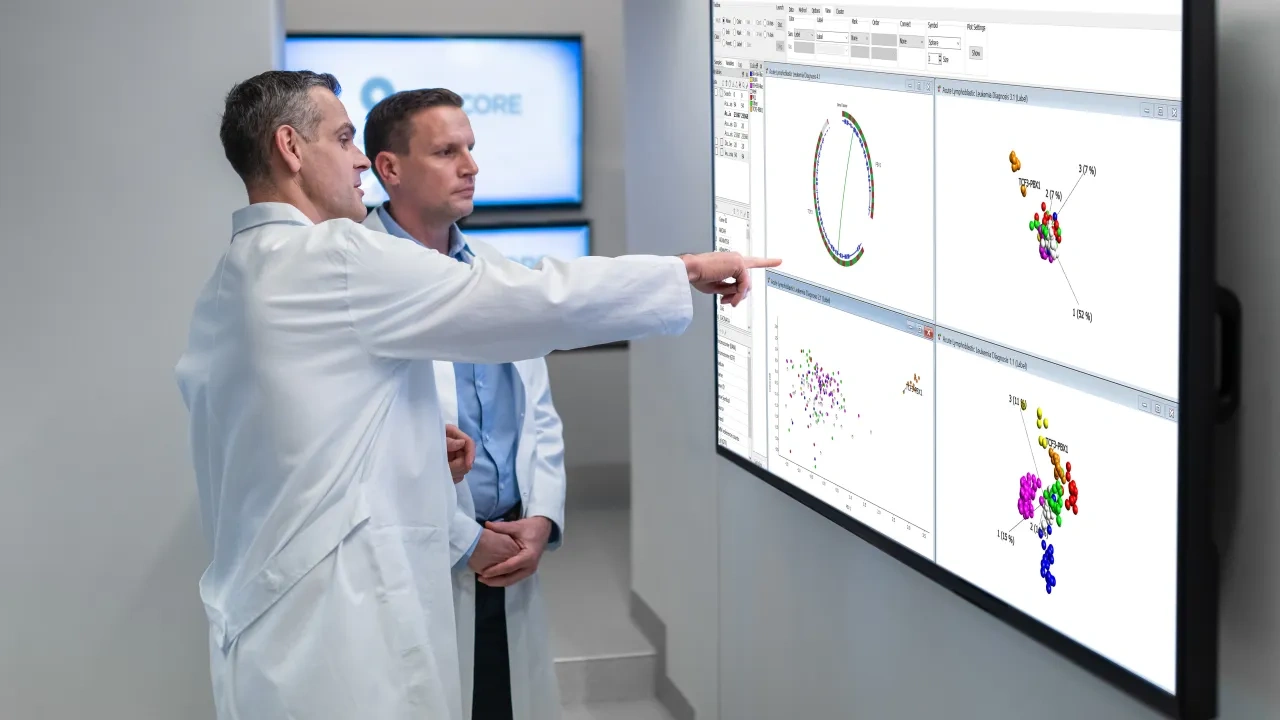
Qlucore Omics Explorer is the powerful visualization-based data bioinformatics analysis tool with inbuilt powerful statistics that delivers immediate results and provides instant exploration and visualization. The program supports in addition to metabolomics data a broad spectrum of multi-omics and NGS data.
Analyze metabolomics data
Metabolomics refers to the systematic identification and quantification of the small molecule metabolic products (the metabolome) of a biological system (cell, tissue, organ, biological fluid, or organism) at a specific point in time. Mass spectrometry and NMR spectroscopy are the techniques most often used for metabolome profiling.
Qlucore Omics Explorer lets the researcher freely examine and analyze data from metabolomics experiments.
Key functionalities:
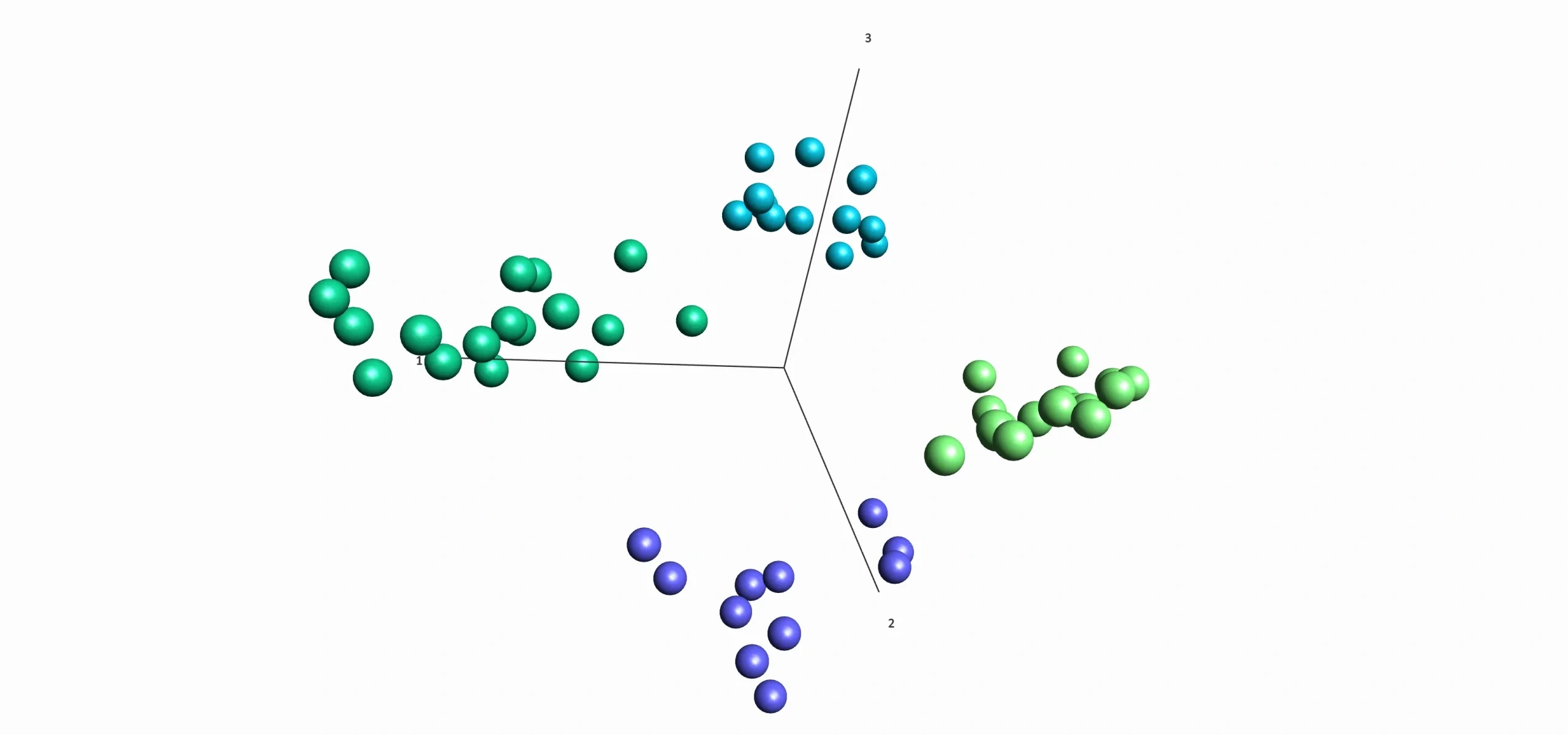
- Investigate any structure in data using variance filtering combined with PCA.
- Perform statistical filtering using for instance ANOVA to enhance results.
- Generate a list of metabolites that classifies data based on a selection of statistical tests: F-test, t-tests or regression.
- Visualize results in any of the 14 supported plot types.
- Export lists for further analysis, such as enrichment and KEGG pathway analysis.
- Use hierarchical clustering or PCA to indentify subgroups.
- Work with Variable PCA plots to find correlation and networks among selected metabolites.
- Export data and plots with a one-button press
Does it work on my data?
Answer the four quick questions below and find out if you can use Qlucore on your data.
For more details about supported data formats and data import see Data Import or Contact us with questions.
Case studies
RNA-Seq analysis using Qlucore
Stanford University, USPerforming gene expression analysis based on RNA sequencing data, in Dilated Cardiomyopathy studies.