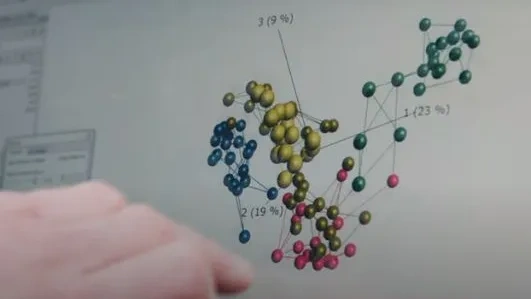Explore
Instant exploration is one of the key features in Qlucore Omics Explorer tailored to generate actionable results. As a user, you decide your own workflow and exploration starting point, if it is a PCA or UMAP plot to check for outliers and batch effects or if it is through variance fitlering to investigate signals in the data. Use the extensive range of plots and statistical filters to explore your multi-omics and NGS data instantly. All through the exploration you are in control and can define the steps to meet your specific needs. Through the Log and status window you keep track of the steps taken.
Explore and find patterns in your omics data
To drive new results, the program puts the user in control of data exploration.
To enhance the results and support cutting edge research a number of functions are supported.
- Extensive range of plots and statistical filters and methods that work in harmony.
- Work with multiple statistical filters optimized for different types of data, examples being DESeq2 tests and ANOVA.
- Use the Biomarker Workbench to identify biomarkers.
- Compare your own results with public dataset such as published at GEO and TCGA.
- Work with multiple data sets.
- Visualize multiple annotations at the same time both in 2D and 3D plots such as PCA using a combination of colors and symbols.
- Extensive coloring options. Data can be colored according to clinical information as well as expression level, p-value and q-value.
- Selections. Work with different parts of the data.
- Explore NGS data in the fast Genome browser that Supports dynamic filtering on a wide range of entities.
- Use k-means clustering, PCA, t-SNE, UMAP and hierarchical clustering to generate hypotheses and do datamining.
- Compare your own results with data published at GEO and TCGA.
- Count data is supported.
Read about all features here.