NGS software
Qlucore Omics Explorer is a powerful visualization-based data analysis tool with inbuilt powerful statistics that delivers immediate results and provides instant exploration and visualization. The program supports a broad spectrum of NGS and Omics data. With the NGS module the genomic analysis is enabled.
Analyzing NGS data
Next-generation sequencing (NGS) technologies transforms among other research areas, cancer research by advancing our understanding of cancer pathogenesis and significantly improving the detection, diagnosis, and treatment of these diseases.
The program offers a broad suite of options for Next Generation Sequencing (NGS) data to analyze and what type of methods you can apply. The NGS module is an add on the Qlucore Omics Explorer base module and all base module functionality is available.
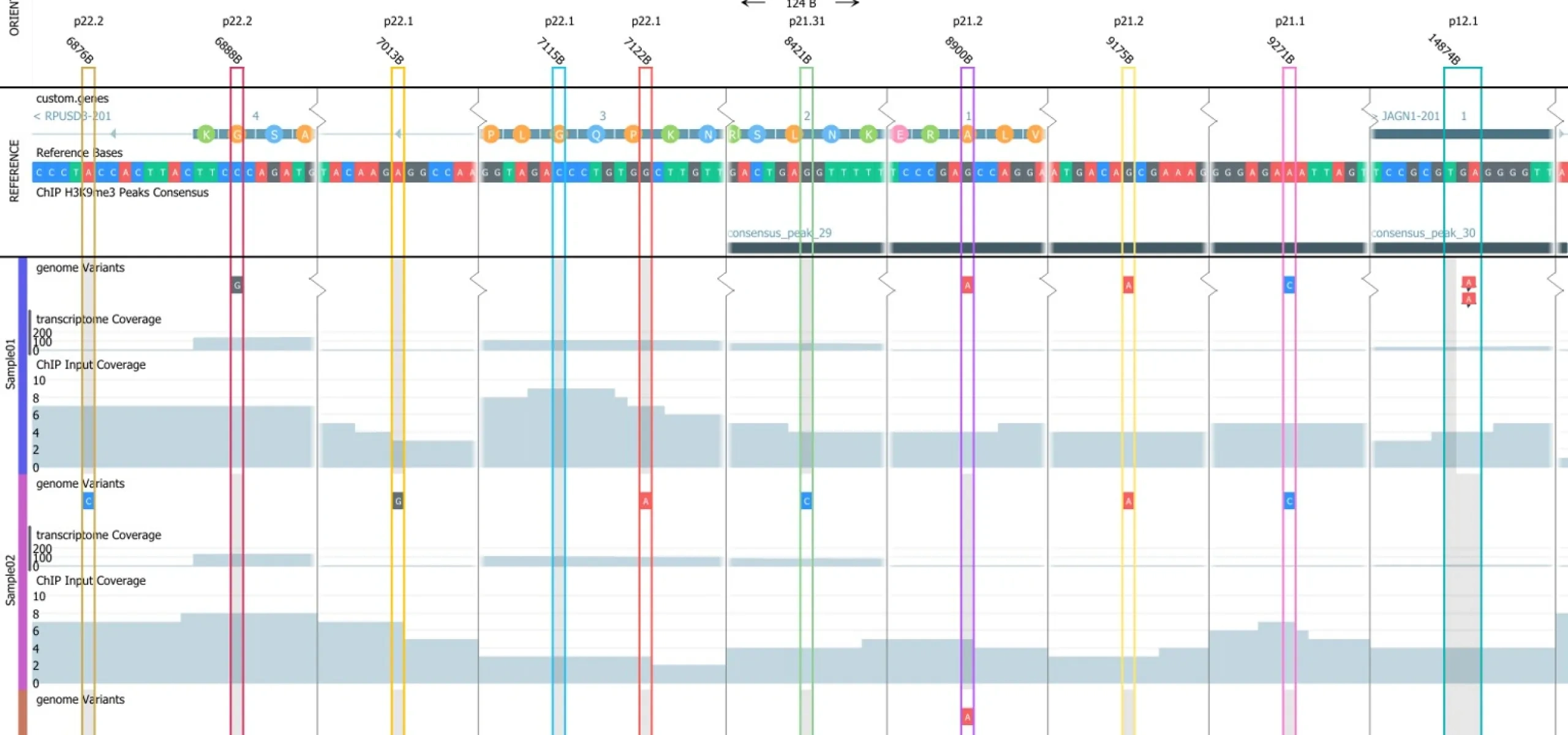
In the center for the NGS data analysis is the the highly interactive and flexible Genome Browser with unique dynamic filtering options. Some examples on available analysis options are:
- ExploreRNA-seq data for deeper insights with the synchronized approach, using both expression levels and genomic information. Note that expression based RNA-seq analysis can be done with the base module.
- variants with the inbuilt variant caller and visualize content after applying filters on quality and/or type.
- Use external variant data base content to enhance visualization and filter against. Import standard vcf files.
- Perform analysis on ChIP-seq and ATAC-seq data.
- Visualize your NGS data in the Genome Browser in a easy to use environment and control what you see using standard user interface controls – no scripting.
- Filter and select based on the content of standard files such as GTF and BED. Freely select and deselect sample groups.
- Easy to use filtering using check boxes and sliders which regions to include or exclude. Find and identify variants with a few key-presses.
- Analyze regions with a Read Coverage above a slider-defined cutoff or make selections based on the built in variant caller.
- Analyze detected gene fusions using the Gene Fusion Workbench. Examine fusions with the Genome Browser such as directional information of each of the partner genes and the full sequence including the breakpoint. Visualize gene fusions with circle plots.
The innovative pre-processing, which only takes place once, makes the performance requirements manageable and the NGS module is easy to deploy organizational-wide*. Full interactivity is provided for few or hundreds of samples, both for RNA-seq and DNA-seq data. The module accepts standard file formats such as FASTA, BAM, VCF, GTF and BED and results can be exported as reports or high quality images and reports.
Learn more
NGS module from a Biologist point of view
Example: Analyzing a yeast data set. Next generation sequencing (NGS) is one of the most powerful technologies we have to gather data on biological samples. While it is used to determine genetic content and to assay RNA abundance, it is also used in combination with diverse protocols to investigate DNA/RNA binding proteins or to assess the composition of non-clonal or metagenomic populations.
Qlucore Omics Explorer NGS Module feature overview
Qlucore Omics Explorer (QOE) supports the user with fast, simple and visual analysis of
measured data. The NGS Module is an add-on module that will enable additional functionality
related to data generated with NGS technologies and will make it possible to interactively and
dynamically analyze and explore NGS data both from DNA and RNA.
Does it work on my data?
Answer the four quick questions below and find out if you can use Qlucore on your data.
For more details about supported data formats and data import see Data Import or Contact us with questions.
Case studies
RNA-Seq analysis using Qlucore
Stanford University, USPerforming gene expression analysis based on RNA sequencing data, in Dilated Cardiomyopathy studies.
Analysis of public data using Qlucore
Beijing Normal University, ChinaThis case study is an example of how the use of public information from multiple sources was used to propose a new classification for glioma cancer.
Using Qlucore in epigenetics research studies
Cancer Genetics Program at the Hospital for Sick Children (SickKids) in Toronto, Canada.A range of samples including DNA from patient blood, primary tissue from tumors, and cell lines, are studied.