ChIP-seq and ATAC-seq analysis
Easy and flexible bioinformatics software for ChIP-seq and ATAC-seq data analysis.
Qlucore Omics Explorer allows for comprehensive analysis of peak data such as ChIP-seq and ATAC-seq. The program includes full work-flow support - from normalization to data export. The main components of the peak analysis are all included and easy accessible. The processing includes
- Peak detection
- Consensus peaks
- Count matrix generation.
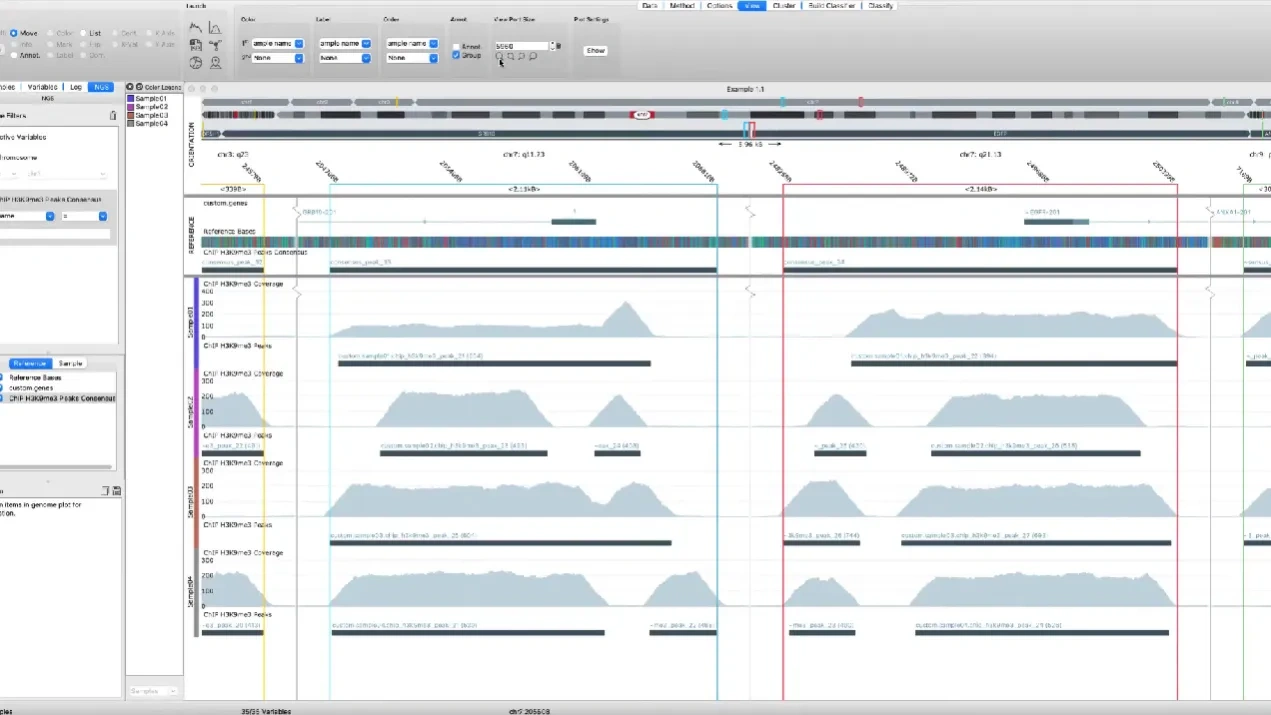
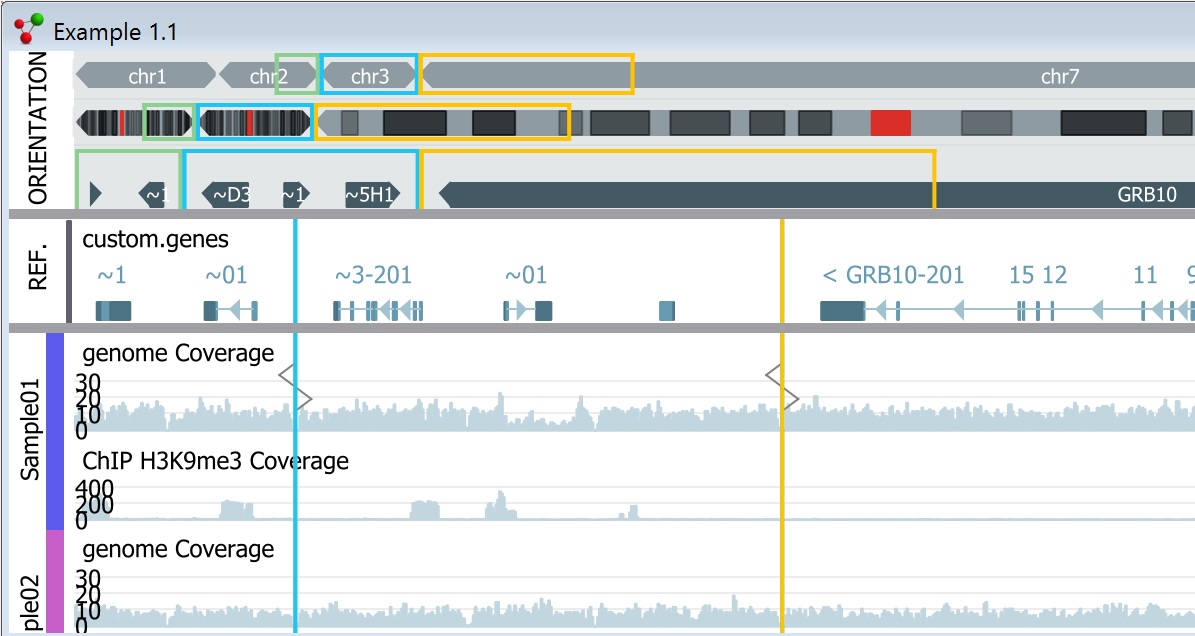
The included Genome Browser includes flexible visualizations and powerful filtering options, making it easy to find interesting features.
Analyze ChIP-seq and ATAC-seq data
The peak analysis support in Qlucore Omics Explorer allows for comprehensive analysis of peak data such as ChIP-seq and ATAC-seq. The main components of the peak analysis processing are peak detection, consensus peaks and count matrix generation. The genome browser in the NGS module includes powerful filtering options for the peak analysis, making it easy to find interesting features. The browser allows the user to annotate peaks and export the results as a bed file. If the experiment also includes RNA-seq data, it is possible to generate a count matrix for RNA-seq and swap between the RNA-seq and ChIP/ATAC-seq count matrices during the analysis.
Key functionalities
- Full work-flow support. From normalization to data export.
- Peak detection and visualization of detected peaks in dedicated tracks.
- Peak variable list with peak information automatically created.
- Visualizations (Genome browser, line plots, heatmaps, pie chart).
- Use the List tool to select Peaks and add them to a variable list that can be saved.
- Option to save data as a BED file.
The peak analysis support requires Next Generation Sequencing module for Qlucore Omics Explorer. To learn more about NGS module.
Watch video and learn more
"Easy and fast ChIP seq analysis tailored for research biologists"
Webinar
Introduction
Easy and fast ChIP-seq
Performing peak analysis is quick and easy in Qlucore Omics Explorer. With the interactive Genome Browser, you add filters to the ChIP-seq data to qui...
Watch now
Introduction
Interactive Genome Browser in the NGS module
The NGS module in Qlucore Omics Explorer enables exploration and analysis of sequencing data such as DNA-seq&n...
Watch now
Qlucore Omics Explorer NGS Module feature overview
Qlucore Omics Explorer (QOE) supports the user with fast, simple and visual analysis of
measured data. The NGS Module is an add-on module that will enable additional functionality
related to data generated with NGS technologies and will make it possible to interactively and
dynamically analyze and explore NGS data both from DNA and RNA.
NGS module from a Biologist point of view
Example: Analyzing a yeast data set. Next generation sequencing (NGS) is one of the most powerful technologies we have to gather data on biological samples. While it is used to determine genetic content and to assay RNA abundance, it is also used in combination with diverse protocols to investigate DNA/RNA binding proteins or to assess the composition of non-clonal or metagenomic populations.