Gene Fusion Workbench
Fusion genes are established disease drivers in several cancer types. Detection of fusion genes using RNA-seq technologies can be done with several tools and pipelines.
The Gene Fusion Workbench, including the circle plot, enables easy analysis and visualization of detected fusion genes.
Gene Fusion Workbench (Research Use Only)
The Gene Fusion Workbench is a flexible way for analysis of detected fusion genes and it is included both as part of the NGS module in Qlucore Omics Explorer and in Qlucore Insights. Input data from many sources are supported, examples are: Star Fusion, Fusion Catcher, Arriba, Manta and DRAGEN pipelines.
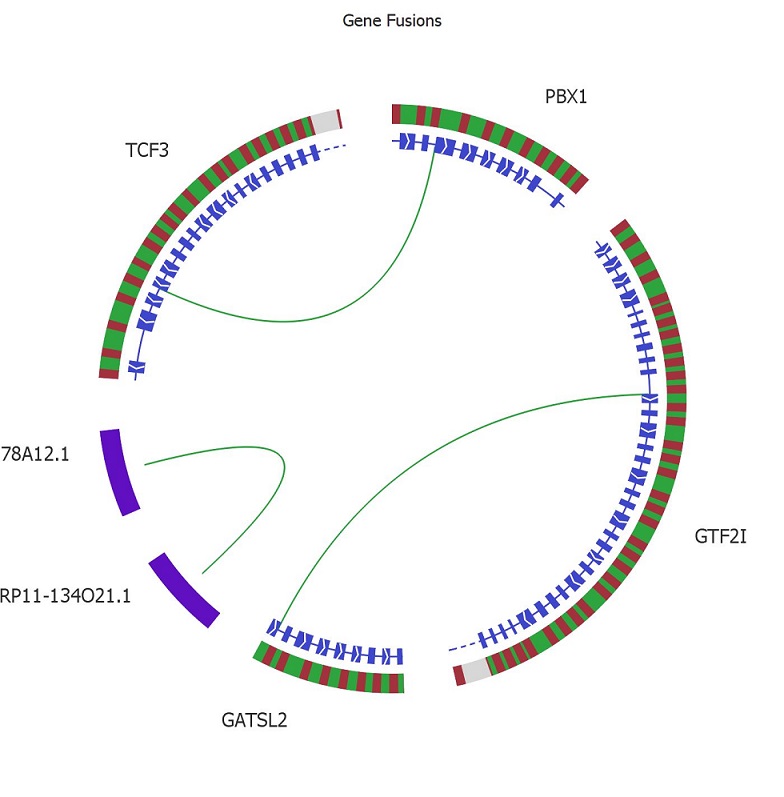
The workbench is shipped with the Mitelman and the Tumor Fusion reference data bases. By combing filters of various types such as presence in the reference data bases and quality, detection of both novel and well-established fusions is straightforward. The fusions can be thoroughly examined in the genome browser and visualized in the Circle plot for a great overview.
Key functionalities:
- Gene fusion discovery. Investigate new and not only known gene fusions.
- Filtering options to quickly sort and navigate the gene fusions.
- Comparison with databases. Each detected gene fusion is compared to Mitelman, TumorFusions databases.
- Well-annotated view of the genetic context including directional information
- Full sequence available including breakpoint.
- Easy look of Gene names and IDs via direct links to NCBI, Ensembl, HGCN and the Mitelman database.
The Workbench includes support for output files from STAR-Fusion, FusionCatcher, Arriba, DRAGEN and Manta (Illumina’s TruSight Oncology 500 panel) gene fusion callers
The Gene Fusion Workbench is included in Qlucore Insights. In Qlucore Omics Explorer, the Gene Fusion Workbench requires the Next Generation Sequencing (NGS) module add-on.
To learn more about NGS module.
